Imagine trying to navigate an unfamiliar city but you’ve lost your smartphone. You don’t have a map. And the street signs are in a foreign language. This is how neuroscientists feel about studying the brain.
Billions of neurons in the human brain make trillions of connections with each other, forming an intricate maze of roads (neural pathways) that make us who we are. A neural pathway is a series of connected neurons that send signals from one part of the brain to another. These networks govern how we think, store our memories and interact with each other. Neuroscientists don’t yet have the technology to outline even a small fraction of the connections within the human brain. We have no map.
But an international team of scientists at the MRC Laboratory of Molecular Biology, University of Cambridge, and Johns Hopkins University has made a breakthrough. We mapped the most complex brain to date: an insect. This brain is in many ways similar to the human brain, with two hemispheres, a brainstem-like structure and a spinal cord equivalent that controls the animal’s muscles.
Understanding this brain may give us many clues about how the human brain works.
There are only a few brain maps, or connectomes, available today. Those that exist are for simple animals, such as worms or sea squirts, which have a few hundred neurons.
You need expensive equipment to map brains, such as powerful electron microscopes to image the tiny synapses between neurons, each measuring roughly 10 billionth of a metre. Synapses are junctions where neurons communicate with each other. Even for simple brains, it takes years to reconstruct neural roadways, synapses and neurons.
In a recent study published in Science, my co-authors and I mapped a fruit fly larva (Drosophila melanogaster) brain with 3,016 neurons and more than half a million synaptic sites. These insects are surprisingly complex. Fruit fly larvae can feel pain, form good and bad memories, and work in a team with their neighbours to forage for food.
What we learned
The first thing we discovered was that fruit fly neurons communicate with each other in not one, but four different ways.
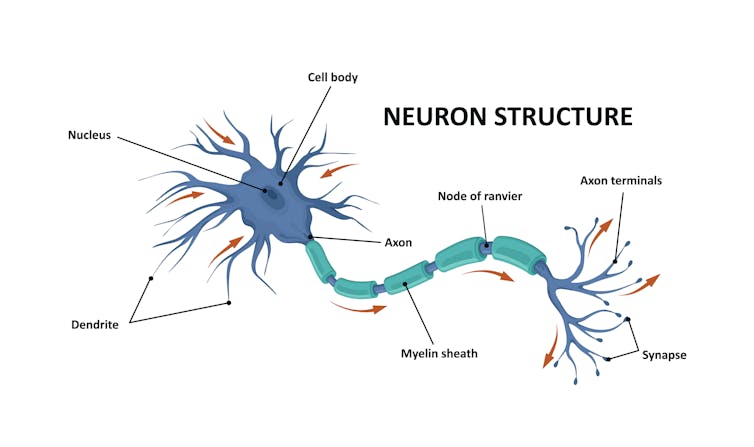
Scientists believed the main way neurons communicated with each other was using its cable (axon) to send neurotransmitters (chemical messengers) to the dendrite of another neuron. Dendrites are appendages designed to receive signals from other cells. However, this was only true for half of connections in the fruit fly larva brain. Sometimes, fruit fly neurons send messages from axon-to-axon, dendrite-to-dendrite, or dendrite-to-axon.
Researchers were aware of these kinds of connections, but little was known about them or how many there were. Our team was shocked by the sheer number of them, which accounted for nearly half the brain’s connectivity. They allowed neurons to connect with each other in ways we didn’t consider before, for example, by combining multiple connection types simultaneously.
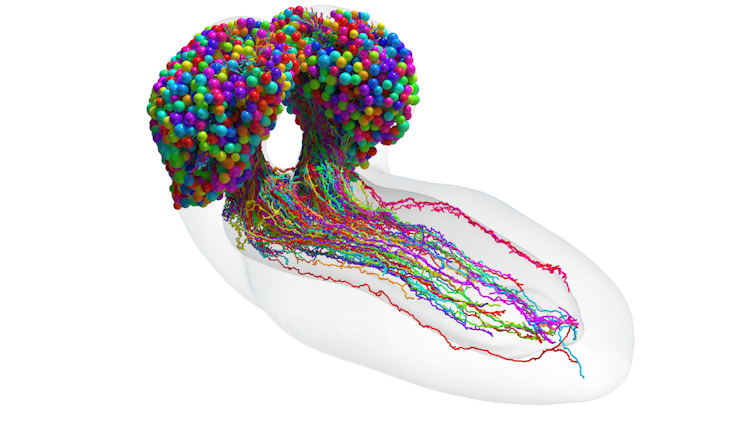
Because there are so many neurons, it’s difficult to think about the brain as one mass of interconnected neurons. So neuroscientists simplify things by grouping together similar “cell types”. Each neuron has a unique shape and role in the brain — essentially its own fingerprint. But there is no consensus about how to define these cell types.
My team stumbled across a new way of breaking down brain structure into its simplest form, using the connections between neurons alone. We found 93 cell types across the fruit fly larva brain. Neurons within each of these groups had similar shapes and performed similar roles in the brain, for example, receiving visual information from the eye.
Next, we focused on the core of the brain in more detail. We developed an algorithm that tracks signals across the brain. Scientists thought different neurons were dedicated to different senses and these signals only later came together. But, using our algorithm, we discovered most neurons multitask, performing different roles depending on the particular sensory cues the fruit fly larva is experiencing. Neurons that processed only one sensory signal were rare.
Algorithms and brains
The fruit fly larva brain showed astonishing similarities with artificial intelligence (AI). Pathways through the brain were layered on top of each other like ResNets, (artificial neural networks that allow deep networks to better learn tasks), allowing it to pack more power. Many brain pathways contained loops. This was shown particularly in the learning and memory centre, perhaps functioning like a recurrent neural network (artificial neural networks with looping connections).
But even simple brains can perform many tasks well, while AI can only do a good job of one task. Plus, brains are much more efficient, using far less energy than AI. If a brain used enough energy to power a lightbulb to solve a problem, AI might need the energy required to build and drive five cars for 120,000 miles to figure out the same puzzle.
AI is becoming more commonplace, with breakthroughs like ChatGPT. This technology is becoming increasingly energy hungry, with the potential to worsen the current climate crisis. Learning how brains can solve complex problems with minimal energy use will be essential to reduce the carbon footprint of AI.
My team’s study is a milestone in understanding the structures of brains. Because fruit flies share many genes with humans, our fruit fly larva brain map may help scientists learn how disease-causing mutations change brain wiring in neurological conditions, such as autism spectrum disorders and schizophrenia.
With a brain map in hand, answers to questions about brain function and dysfunction are within reach. All we need to do is keep exploring.
![]()
Michael Winding is currently affiliated with the Francis Crick Institute, but was a Research Associate at three institutes when this research was conducted: the University of Cambridge, Department of Zoology; the MRC Laboratory of Molecular Biology; and HHMI Janelia Research Campus